Least Squares
How to fit a nonlinear function to data
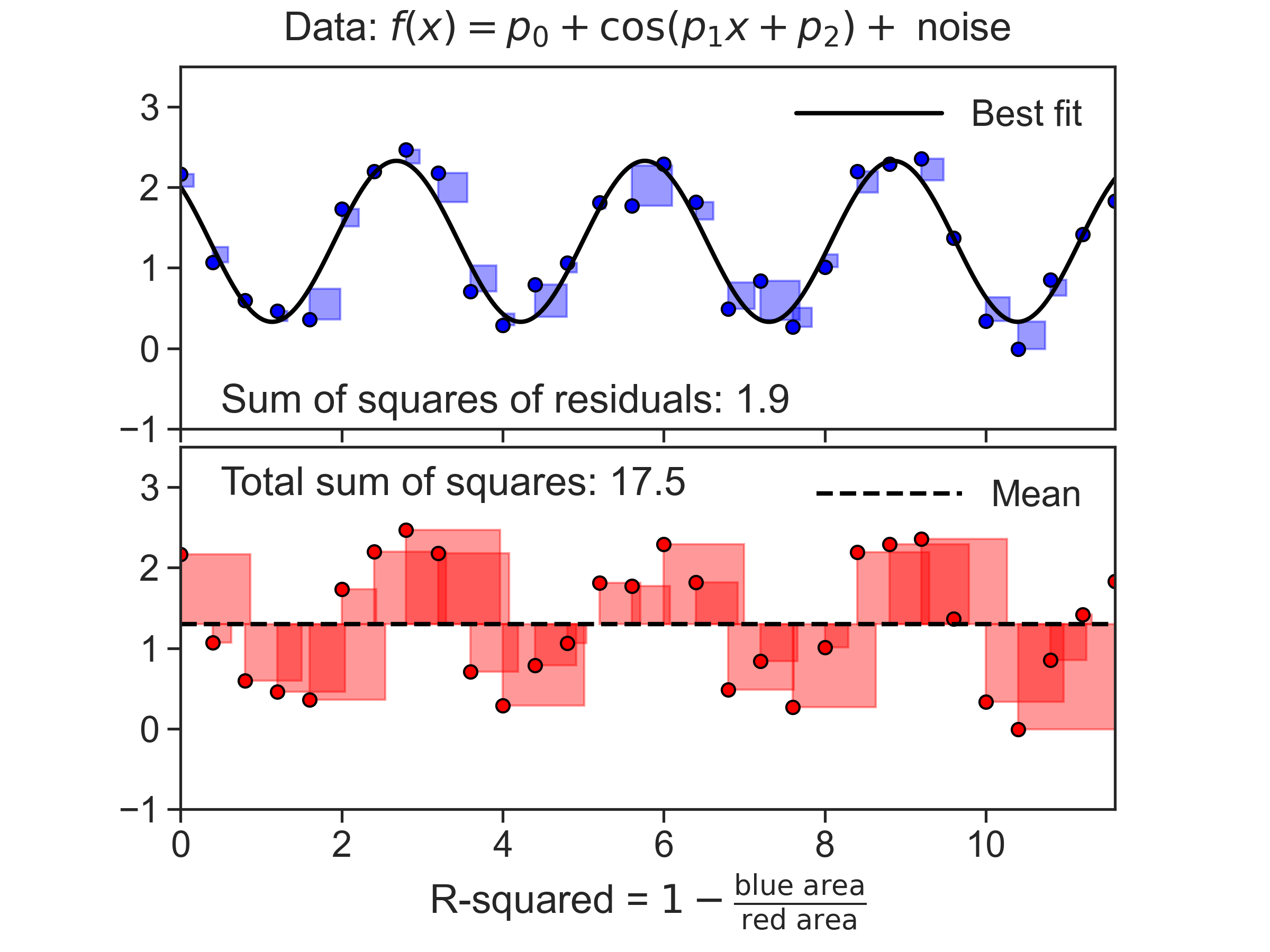
Introduction
This code produces the figure above. It’s main tool is the curve_fit method, that allows us to fit any function to data, and get optimal parameter values.
The code
Define some functions
Now let’s plot some stuff
%matplotlib notebook
fig, [ax1, ax2] = plt.subplots(2, 1, figsize=(8,6), sharex=True, sharey=True,
subplot_kw={'aspect':'equal'}
)
fig.subplots_adjust(left=0.04, right=0.98, top=0.93, bottom=0.15,
hspace=0.05, wspace=0.02)
params = {#'backend': 'ps',
'font.family': 'serif',
'font.serif': ['Computer Modern Roman'],
'ps.usedistiller': 'xpdf',
'text.usetex': True,
# include here any neede package for latex
'text.latex.preamble': [r'\usepackage{amsmath}'],
}
# the parameter values
par = (1.3, 2, 1)
# generating data with noise
y = func(x, *par) + (np.random.random(len(x)) - 0.5)
ax1.plot(x, y, marker='o', ls='None', markerfacecolor="blue", markeredgecolor="black")
ax2.plot(x, y, marker='o', ls='None', markerfacecolor="red", markeredgecolor="black")
# best fit
popt, pcov = curve_fit(func, x, y, p0=(1.5, 1.5, 2.5)) # p0 = initial guess
p0, p1, p2 = popt
# The total sum of squares (proportional to the variance of the data)
SStot = ((y - y.mean()) ** 2).sum()
# The sum of squares of residuals
SSres = ((y - func(x, p0, p1, p2)) ** 2).sum()
Rsquared = 1 - SSres / SStot
# plot best fit
h = np.linspace(x.min(), x.max(), 1001)
fit, = ax1.plot(h, func(h, p0, p1, p2), color='black', linewidth=2)
ax1.legend([fit], ["Best fit"], loc="upper right",
frameon=False, handlelength=4)
# plot mean
mean, = ax2.plot(h, h * 0 + np.mean(y), ls='--', color='black', linewidth=2)
ax2.legend([mean], ["Mean"], loc="upper right", frameon=False, handlelength=4)
# plot blue and red squares
for ind in np.arange(len(x)):
x0 = x[ind]
y0 = y[ind]
# print(x0,y0)
v1 = y0 - func(x0, p0, p1, p2)
v2 = y0 - y.mean()
add_rec(ax1, (x0, y0), -v1, "blue")
add_rec(ax2, (x0, y0), -v2, "red")
ax2.text(0.5, 2.9, r"Total sum of squares: {:.1f}".format(SStot))
ax1.text(0.5, -0.8, r"Sum of squares of residuals: {:.1f}".format(SSres))
# ax2.set_xlabel(
# r"R-squared = $1 - \frac{\text{blue area}}{\text{red area}}$ = " +
# "{:.2f}".format(Rsquared))
ax2.set_xlabel(
r"R-squared = $1 - \frac{\mathrm{blue\ area}}{\mathrm{red\ area}}$"
)
ax1.set_xlabel(
r"Data: $f(x) = p_0 + \cos(p_1 x + p_2)+ $ noise ",
labelpad=12
)
ax1.xaxis.set_label_position("top")
ax1.set(xlim=[x.min(), x.max()],
ylim=[-1, 3.5],
xticklabels=[],
yticks=np.arange(-1, 4)
)
ax2.set(
xticklabels=np.arange(0,13,2)
)
fig.savefig("least-squares.png", dpi=300)/Users/yairmau/opt/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:70: UserWarning: FixedFormatter should only be used together with FixedLocator